



- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource
- …
- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource



- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource
- …
- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource

Maize
(Zea mays Linn.)
To support maize research and breeding, we offer a portfolio of GBTS panels with multiple marker densities. These panels address diverse genotyping needs and enable flexible applications in genetic analysis and selection, providing reliable support for both research and breeding improvement.
For projects with specific genotyping requirements, custom GBTS panels can be developed. Contact us to discuss a solution tailored to your breeding program.
Genotyping by Targeted Sequencing (GBTS) Panels - Ready to use*
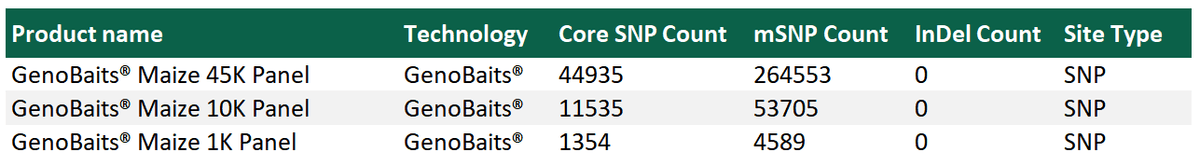
Reference genome: AGPv3
*A ready-to-use GBTS panel is fully developed and assembled, allowing you to submit DNA or leaf tissue samples directly for rapid genotyping and data delivery. All panels can be customized with additional markers for specific traits or loci. To include your loci of interest, please contact us.
- Product Highlight

Broad Polymorphism Coverage
The markers on these panels were selected from Hap-Map3 which is based on the whole genome sequencing of 1,218 major maize inbreds and improved worldwide cultivar accessions.

Marker Density Flexibility
Adjunct SNPs are captured with the target SNPs and form multiple SNP clusters, which are essential for haplotype mapping and structure analysis. In the maize 45K SNP panel, Molbreeding designed total of 264,553 multiple SNP clusters, mSNPs. The Maize 10K panel has 53,705 mSNP and the 1K SNP has total 4,589 mSNP. Different application can select the appropriate marker density to minimize cost and analysis time.

Sample Type Flexibility
Supports diverse sample inputs, including genomic DNA, seeds, dried leaf samples, and plant tissues.
- Application
For Discovery Service
Genetic map construction
QTL analysis
Genome-wide association study
For Breeding Service
Germplasm characterization
Molecular marker-assisted selection
Genome-wide selection breeding
Variety protection, Varietiy authentication
- Report Visualization

GenoBaits® Maize 45K Panel
The markers on this panel were selected from Hap-Map3 which is based on the whole genome sequencing of 1,218 major maize inbreds and improved worldwide cultivar accessions.

GenoBaits® Maize 10K Panel
The10K SNP panel designed using a total of 53,705 multiple SNP capture sites, with a total of 11,535 target SNPs evenly distributed across the 10 maize chromosomes.

GenoBaits® Maize 1K Panel
This 1K Panel designed using a total of 4,589 multiple SNP capture sites, with a total of 1,354 target SNPs evenly distributed across the 10 maize chromosomes.
The duplicate samples consistency is all above 99.41%.
- Publications
- Gao J, Wang S, Zhou Z, et al. Linkage mapping and genome-wide association reveal candidate genes conferring thermotolerance of seed-set in maize. J Exp Bot. 2019;70(18):4849-4864.
- Guo Z, Wang H, Tao J, et al. Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize. Mol Breeding. 2019;39(37).
- Liu HJ, Jian L, Xu J, et al. High-Throughput CRISPR/Cas9 Mutagenesis Streamlines Trait Gene Identification in Maize. Plant Cell. 2020;32(5):1397-1413. DOI: 10.1105/tpc.19.00934
- Wen J, Shen Y, Xing Y, et al. QTL Mapping of Fusarium Ear Rot Resistance in Maize. Plant Dis. 2021;105(3):558-565.
- Guo Z, Yang Q, Huang F, et al. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Commun. 2021;2(6):100230.
- Han L, Jiang C, Zhang W, et al. Morphological Characterization and Transcriptome Analysis of New Dwarf and Narrow-Leaf (dnl2) Mutant in Maize. Int J Mol Sci. 2022;23(2):795.
- Zhang X, Wang M, Zhang C, et al. Genetic dissection of QTLs for starch content in four maize DH populations. Front Plant Sci. 2022;13:950664.
- Huang J, Li Y, Ma Y, et al. The rhizospheric microbiome becomes more diverse with maize domestication and genetic improvement. J Integr Agric. 2022;4:1188-1202.
- Liu R, Cui Y, Kong L, et al. Evaluating the Genetic Background Effect on Dissecting the Genetic Basis of Kernel Traits in Reciprocal Maize Introgression Lines. Genes (Basel). 2023;14(5):1044.
- Gao J, Feng P, Zhang J, et al. Enhancing maize's nitrogen-fixing potential through ZmSBT3, a gene suppressing mucilage secretion. J Integr Plant Biol. 2023;65(12):2645-2659.
- Yu G, Cui Y, Jiao Y, et al. Comparison of sequencing-based and array-based genotyping platforms for genomic prediction of maize hybrid performance. Crop J. 2023;11(2):490-498.
- Luo P, Wang H, Ni Z, et al. Genomic prediction of yield performance among single-cross maize hybrids using a partial diallel cross design. Crop J. 2023;11(6):1884-92.
- Xu F, Liu S, Zhao A, et al. iFLAS: positive-unlabeled learning facilitates full-length transcriptome-based identification and functional exploration of alternatively spliced isoforms in maize. New Phytol. 2024;241(6):2606-2620.
Documents
- If you require marker list or any additional information, please contact us.
Sample Preparation and Submission Guidelines
Brochure

Subscribe to our newsletter
Subscribe now to our regularly released newsletter and be informed about the latest products and special offers.









