- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource
- …
- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource



- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource
- …
- Product
- GBTS Panels
- Software System
- Reagent
- Instrument
- Resource

Pepper
(Capsicum annuum Linn.)
To support pepper (Capsicum spp.) research and breeding, we offer ready-to-use GBTS panels designed for high-throughput genotyping. Developed using diverse pepper germplasm, these GBTS panels provide robust genomic coverage for applications such as genetic analysis, trait discovery, and genomic selection.
For projects with specific genotyping requirements, custom GBTS panels can be developed. Contact us to discuss a solution tailored to your breeding program.
Genotyping by Targeted Sequencing (GBTS) Panels - Ready to use*

Reference Genome: Capsicu_annuum_Ca_59_1.0
*A ready-to-use GBTS panel is fully developed and assembled, allowing you to submit DNA or leaf tissue samples directly for rapid genotyping and data delivery. All panels can be customized with additional markers for specific traits or loci. To include your loci of interest, please contact us.
- Product Highlight

Enriched polymorphism selection and a broad spectrum of variation
Our panels offer comprehensive coverage of diverse genetic variations across germplasm resources, utilizing variation data from 2,361 pepper germplasm samples spanning five widely cultivated pepper species.

Various density designed for different applications
Our ready-to-use pepper GBTS panels are designed with various, flexibly adjustable marker densities, from high to low, to cater to your diverse application scenarios, truly optimizing time and cost. We offer flexible marker number adjustment and upgrade services, ensuring your research remains at the forefront.
- Application
Germplasm Identification and DNA fingerprint construction
Marker-Assisted Backcross Selection
Variety Identification and Parental Line Purification
Genetic Diversity Analysis
Genome-wide selection breeding
Seed Purity Identification
Functional Gene Mining and Identification
(GWAS/QTL/BSA)
- Report Visualization
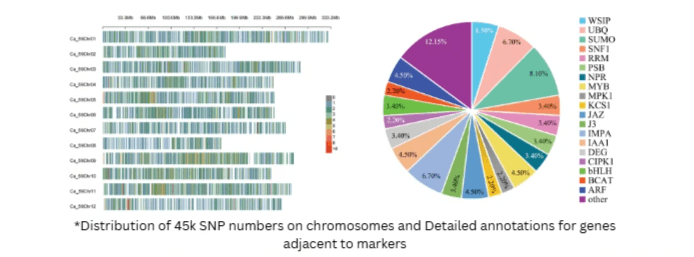
GenoBaits® Pepper 45K Panel
The 45K panel offers high resolution, featuring 44,646 target SNPs along with an additional 27,000 mSNPs (flanking SNPs identified on fragments captured by probes). This panel includes 278 trait-associated markers that cover 30 traits across agronomic, quality, pungency, reproductive, abiotic, and biotic categories, facilitating informed breeding decisions.

GenoBaits® Pepper 20K Panel
Based on the 45K pepper marker panel, we has developed a medium-density 20K pepper panel, which includes 20000 target SNPs and additional 10K mSNPs, with 160 trait-associated markers spanning 28 traits.

GenoBaits® Pepper 10K Panel
The 10K panel includes 10000 target SNPs and additional 5K mSNPs, with 120 target markers associated with 24 traits.
- Validation of GenoBaits Pepper 45K Panel
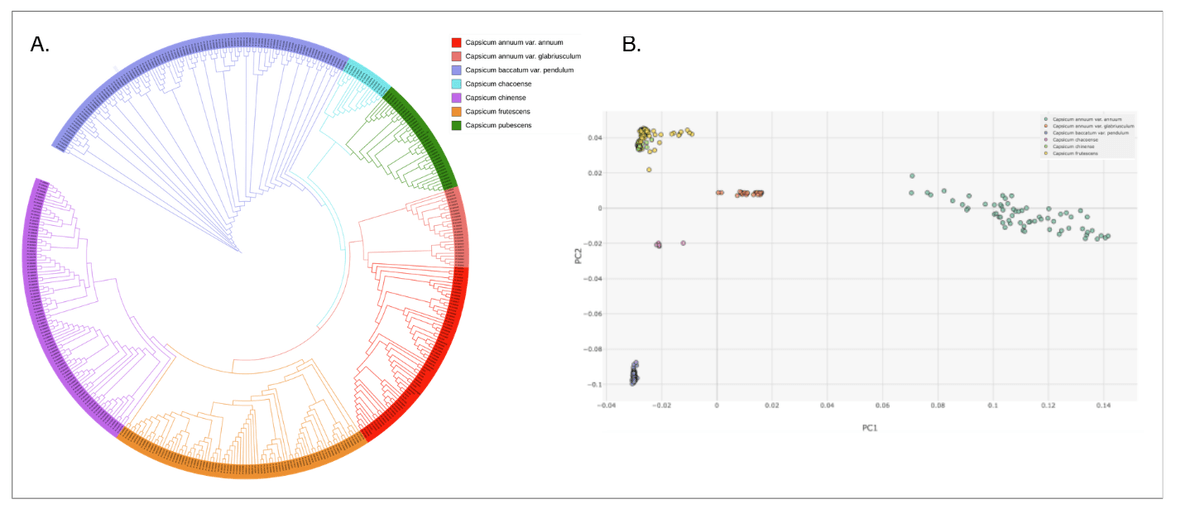
To validate the GenoBaits 45K Pepper Panel, SNP loci are extracted from sequencing data of 436 wild and cultivated accessions across nine Capsicum taxa (PRJNA800056 and PRJNA801499). These samples include diverse cultivars and landraces.
Using the retrieved 45K SNPs, phylogenetic trees and principal component analysis (PCA) are generated to evaluate the panel’s effectiveness in capturing genetic relationships and evolutionary patterns within the Capsicum lineage.
- Published Literatures
- Du H, Yang J, Chen B, et al. Target sequencing reveals genetic diversity, population structure, core-SNP markers, and fruit shape-associated loci in pepper varieties. BMC Plant Biol. 2019;19(1):578. https://doi.org/10.1186/s12870-019-2122-2
- Li Z, Jia Z, Li J, et al. Development of a 45K pepper GBTS liquid-phase gene chip and its application in genome-wide association studies. Front Plant Sci. 2024;15:1405190. Published 2024 Jun 25. doi:10.3389/fpls.2024.1405190
Documents
- If you require marker list or any additional information, please contact us.
Sample preparation and shipping guideline